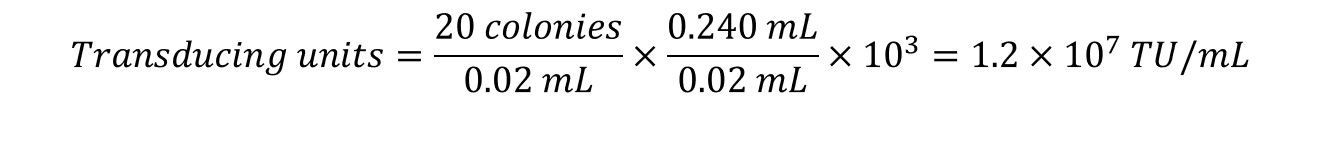Micropanning Assay
Micropanning, also known as a phage capture assay, is used to measure the binding of phage clones to a molecular target. This protocol is typically done after a biopanning (affinity selection) experiment where phage clones of interest have been selected for further analysis. The advantage of the micropanning assay is the relatively high sensitivity compared to traditional ELISA methods of measuring phage biomolecular interactions. The signal measures the titer of infective phage particles; thus, there is no non-phage background that obscures the signal.
This protocol is based on and modified from the The Laboratory of George P. Smith at the University of Missouri. The protocols were previously hosted by http://www.biosci.missouri.edu/smithgp/
Reagents
Note: This protocol describes a micropanning assay using filamentous phage based on the Fd-tet vector (tetracycline resistant) and uses K91BluKan E. coli cells (kanamycin resistant) that are cultured in NZY medium. The protocol can be modified to accommodate other phage variants and E. coli strains taking into account their optimal medium and antibiotic resistance.
- NZY medium
- Terrific broth
- E. coli (K91BluKan)
- Kanamycin
- Tetracycline
- NZY agar
- TBS
- Blocking solution such as 5% BSA in TBS
- Wash buffer (0.5% Tween-20 in TBS)
- TBS gelatin
- Elution buffer (0.1% BSA, 0.1 M HCl using glycine to adjust the pH to 2.2)
- Neutralization buffer (1 M Tris, HCl pH 9.1)
E. coli cultures
1. Inoculate 2 mL of NZY medium with 100 µg/mL kanamycin with K91BluKan E. coli and incubate overnight at 250 rpm at 37ºC.
2. Use 100 µL of the overnight culture to inoculate 10 mL terrific broth in a 125 mL flask. Shake at 250 rpm and 37ºC until the culture gets turbid.
3. Begin reading the OD600 of 1/10 dilutions. When the OD600 of a 1/10 dilution reaches 0.125-0.25, slow the shaking down to 50 rpm to allow sheared F-pili to regenerate. Use these cells for titering within 1 h.
Micropanning
4. Immobilize the antigen (molecular target) on a 96-well plate according to the manufacturer’s instructions.
-Most often the antigen is a recombinant protein, but can also be a carbohydrate, cells, tissue, or other biological or non-biological molecule.
5. Fill each well with blocking solution and incubate for 2 h at room temperature.
-Use a blocking solution according to the manufacturer’s instructions. Alternatively, a blocking solution containing 5% bovine serum albumin (BSA) in Tris-buffered saline (TBS), NaHCO3, or other suitable buffer.
6. Wash each well five times with 0.5% Tween in TBS, preferably using an automated plate washer.
7. Add 50 µL input phage and incubate at 4°C for 2 h on a rocker or at slow shaking.
-Add TBS to some wells to serve as a negative control (no input phage).
-The viral particle concentration of the input phage can be up to ~1010 virions/mL (~5×108 TU/ml). The input phage concentration should be optimized and can be reduced for analysis of high-affinity phage clones.
-For qualitative micropanning to ascertain whether a clone binds strongly to the antigen, an input phage concentration of ~107 virions/mL can be used. Also, the serial dilutions in the titering steps can be skipped (step 8).
8. Wash the wells ten times with 0.5% Tween in TBS.
9. Add 20 µL elution buffer and incubate for 10 min at room temperature.
-1% BSA, 0.1 M HCl using glycine to adjust the pH to 2.2 can be used as an elution buffer. Other suitable buffers with detergent may also be used.
10. Transfer the eluate (output phage) to a corresponding well of a fresh 96-well plate to which 3.75 µL 1 M Tris, HCl pH 9.1 has already been added to increase the pH.
11. In the fresh 96-well plate, make serial 10-fold dilutions by passing 2.2 µL of the neutralized eluate into 20 µL TBS gelatin diluent.
-Note: The input phage can also be tittered to determine an accurate percent yield. It is recommended to dilute the input phage to 105 V/mL in TBS gelatin prior to adding to the 96-well plate.
12. Add 20 µL of E. coli cells from step 3. Incubate at room temperature for 10 min to allow infection.
13. Add 200 µL of NZY medium with 100 µg/mL kanamycin and incubate for 30 min at 37C°.
14. Spot 20 µL from each well in duplicate onto a dish with NZY agar supplemented with 100 µg/mL kanamycin and 40 µg/mL tetracycline. Let the spots dry and incubate the plate upside down at 37°C for 12 h to overnight.
15. Count the colonies in each spot.
16. Calculations:
Protocol
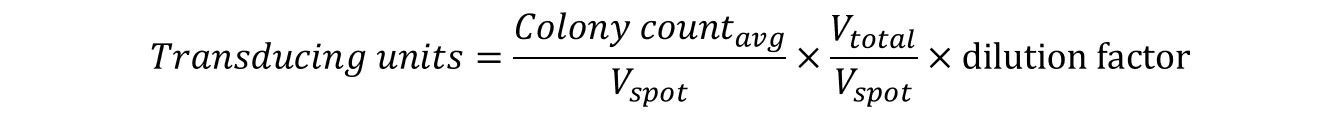
Example: 20 µL from the103 dilution was spotted in duplicate on an agar plate. The colonies from each spot was counted and found to be 18 and 22. The TU/mL was calculated as follows: